Previous
Lecture
Home
Next
Lecture
Table
of Contents

2. To define the mode of action of antibiotics
Specific educational objectives (terms and concepts upon which you will be tested)
Many metabolic activities of the bacterial cell differ significantly from those in the human cell. At least theoretically these differences can be exploited in the development of chemotherapeutic agents. Ideally, an antimicrobial agent should have its maximal effect on the bacterial cell and have little or no effect on the human cell. In reality there is almost always some effect on the human be it induction of hypersensitivity or liver or kidney toxicity. Despite some adverse reactions in the human, effective antibiotics have been developed that have one ore more of these modes of action on the bacterial cell:
A. Inhibition of cell wall synthesis
B. Alteration of cell membranes
C. Inhibition of protein synthesis
D. Inhibition of nucleic acid synthesis
E. Antimetabolic activity or competitive antagonism
Since bacteria have a cell wall made up of repeating units of peptidoglycan and human cells lack this feature, it would seem that the bacterial cell wall presents an ideal target for chemotherapy. Indeed, this has been the case; the following antibiotics have been developed as inhibitors of cell wall synthesis:
A. Beta-lactam antibiotics
|
|
|
|
| Cephalothin | Cefamandole | Cefotaxime |
| Cephaprin | Cefuroxime | Ceftizoxime |
| Cefazolin | Cefonicid | Ceftriaxone |
| Cefelixin * | Ceforanide | Ceftazidime |
| Cephradine * | Cefaclor * | Cefoperaxone |
| Cefadroxil * | Cefoxitin | Moxalactam |
| Cefotetan |
4. Thienamycins
5. ß-lactamase inhibitors (e.g., clavulanic acid)
C. Fosfomycin (Phosphonomycin)
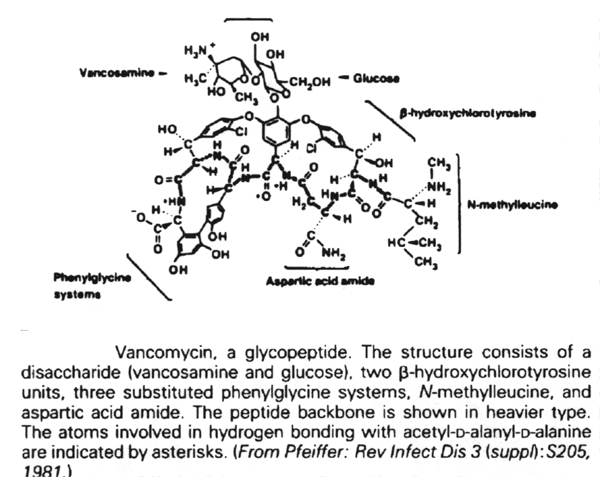
D. Vancomycin
E. Bacitracin
F. Ristocetin
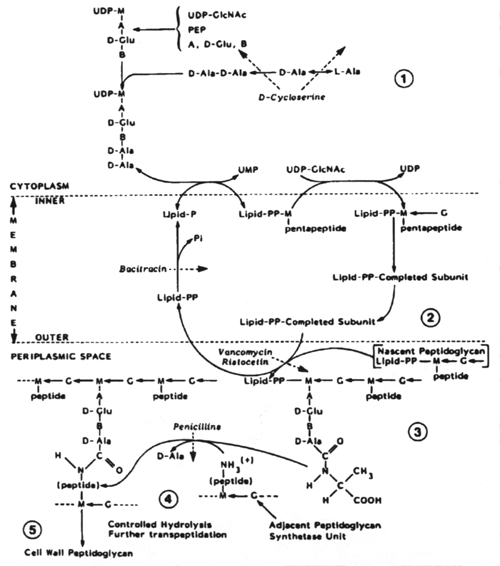
Biosynthesis of peptidoglycan showing the site of action of various cell wall inhibitors. (From Tipper: In Queener, Webber, Queener (eds): Beta-Lactam Antibiotics for Clinical Use. Marcel Dekker, 1986.) From Zinsser Microbiology, 20th ed., Joklik et al., p. 155, Fig. 9-1. Reproduced with permission.
The biosynthesis of peptidoglycan consists of three stages, each of which occurs at a different site in the cell.
Stage 1 occurs in the cytoplasm. In this stage the recurring units of the backbone structure of murein, N-acetylglucosamine and N-acetyl-muramylpentapeptide are synthesized in the form of their uracil diphosphate (UDP) derivatives. The only antibiotic that affects this stage of cell wall metabolism is D-cycloserine. D-cycloserine is a structural analog of D-alanine; it binds to the substrate binding site of two enzymes, thus being extremely effective in preventing D-alanine from being incorporated into the N-acetylmuramylpeptide.
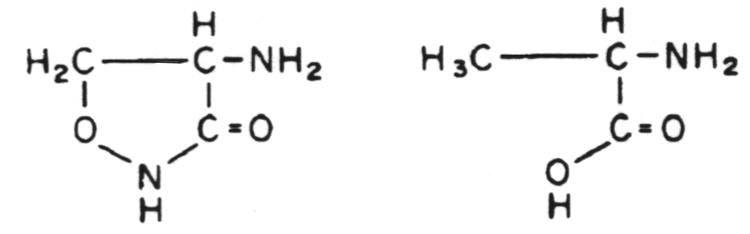
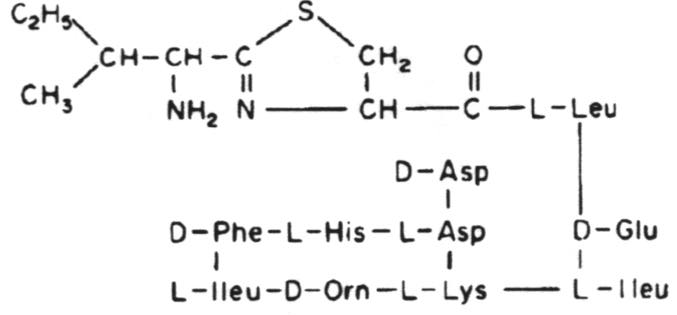
Bacitracin A. One of a group of polypeptide antibiotics containing a thiazoline ring structure.
Stage 3 occurs in the periplasmic space (in gram-negative bacteria) and in the growing peptidoglycan of the cell wall. This is a complex metabolic sequence which offers multiple targets for chemotherapeutic agents. The earliest acting of these are vancomycin and ristocetin. They act by binding to the D-alanyl-D-alanine peptide termini of the nascent peptidoglycan-lipid carrier. This inhibits the enzyme transglycosylase.
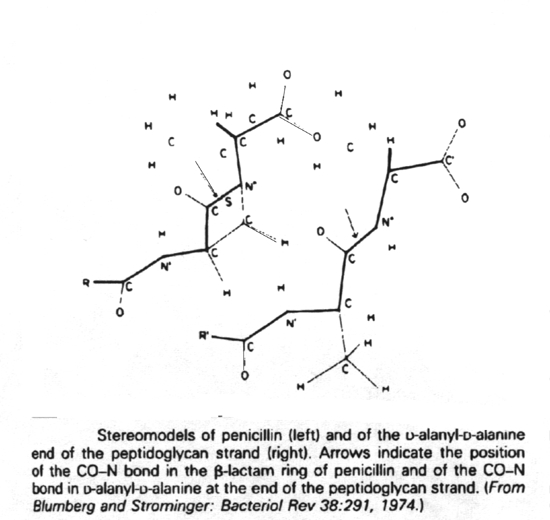
Stage 3 of biosynthesis continues with transpeptidation and the binding of soluble uncrosslinked, nascent peptidoglycan to the preexisting, crosslinked, insoluble cell wall peptidoglycan matrix. The ß-lactam antibiotics are structural analogs of the D-alanyl-D-alanine end of the peptidoglycan strand. In the cell wall there are as many as seven enzymes (depending on the bacterial species) which bind peptidoglycan units via their D-alanyl-D-alanine residues. The ß-lactams fill these substrate binding sites and thus prevent the binding of D-alanyl-D-alanine residues. Enzymes binding beta-lactam antibiotics are known as penicillin-binding proteins.
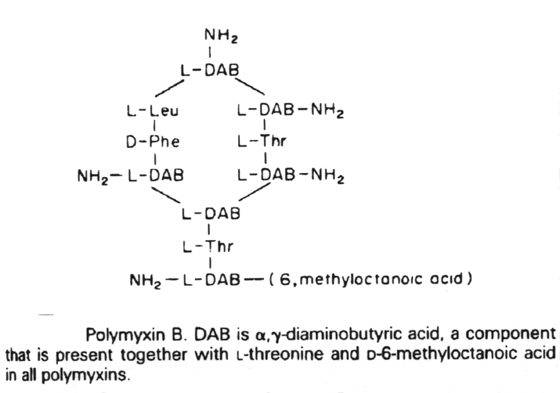
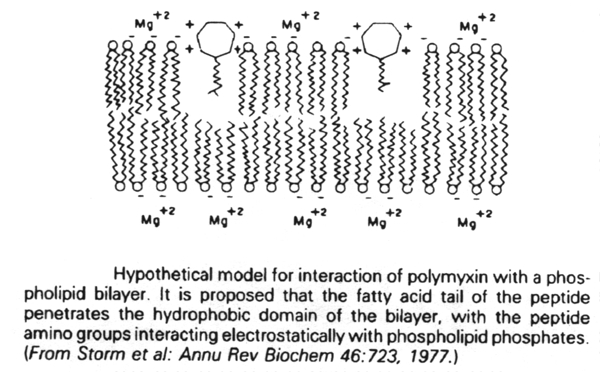
The cytoplasmic membrane of bacteria is only affected by two clinically-used antibiotics. These are polymyxin B and polymyxin E (colistin). They act by competitively replacing Mg2+ and Ca2+ from negatively charged phosphate groups on membrane lipids. The result is disruption of the membrane.
The major group of antibacterial agents that act by blocking DNA synthesis/activity is the quinolone group.
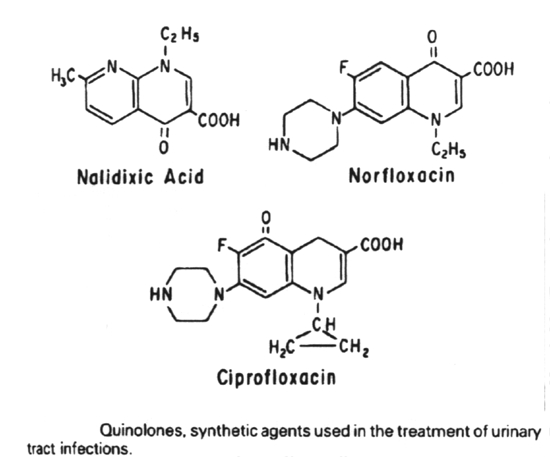
The quinolones all act by blocking the A subunit of DNA gyrase and inducing the formation of a relaxation complex analogue.
DNA gyrase introduces negative superhelical turns into duplex DNA, using the energy of ATP. This is the crucial enzyme that maintains the negative superhelical tension of the bacterial chromosome.
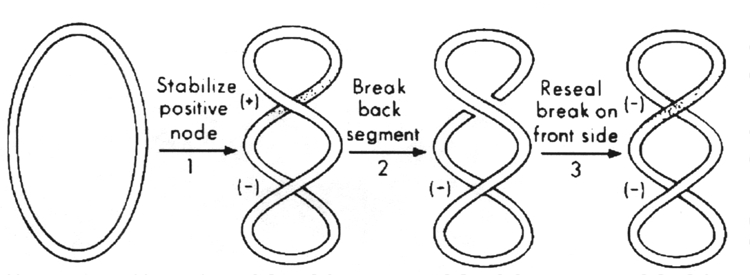
The sign-inversion mechanism for DNA gyrase.
The quinolones include:
norfloxacin, ciprofloxacin - second generation
Protein synthesis is the end result of two major processes, transcription and translation. An antibiotic that inhibits either of these will inhibit protein synthesis.
Transcription
During transcription, the genetic information in DNA is transferred to a complementary sequence of RNA nucleotides by the DNA-dependent RNA polymerase. This enzyme is composed of 5 subunits, ß, ß', a, a' and o. Antibiotics that either alter the structure of the template DNA or inhibit the RNA polymerase will interfere with the synthesis of RNA, and consequently with protein synthesis.
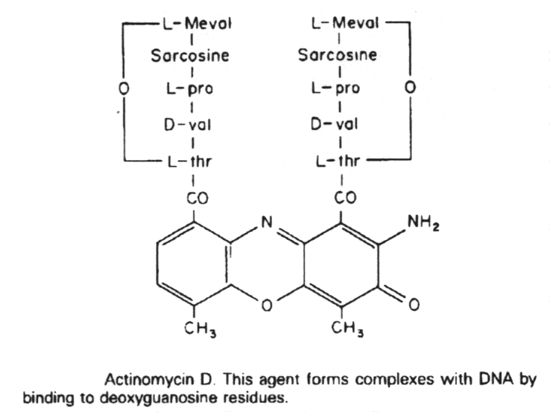
Actinomycin D binds to guanine in DNA, distorting the DNA, and thus blocking transcription.
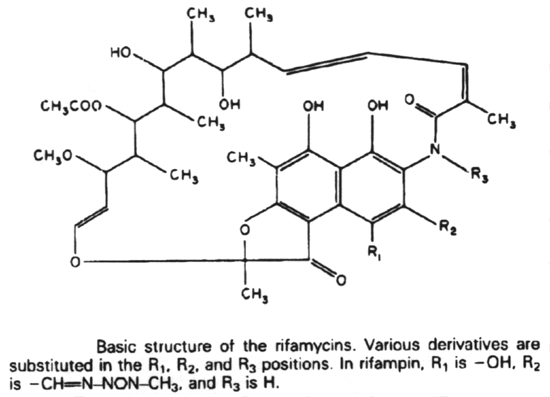
Rifampin (Rifampicin or Rifamycin) inhibits protein synthesis by selective inhibiting the DNA-dependent RNA polymerase. It does this by binding to the ß subunit in a non-covalent fashion.
Translation
In bacterial cells, the translation of mRNA into protein can be divided into three major phases: initiation, elongation, and termination of the peptide chain. Protein synthesis starts with the association of mRNA, a 30S ribosomal subunit, and formyl-methionyl-transfer RNA (fMet-tRNA) to form a 30S initiation complex. The formation of this complex also requires guanosine triphosphate (GTP) and the participation of three protein initiation factors. The codon AUG is the initiation signal in mRNA and is recognized by the anticodon of fMet-tRNA. A 50S ribosomal subunit is subsequently added to form a 70S initiation complex, and the bound GTP is hydrolyzed. In the elongation phase of protein synthesis, amino acids are added one at a time to a growing polypeptide in a sequence dictated by mRNA. It is this phase that is most susceptible to inhibition by a number of antibiotics. For many of these the ribosome is the target site. There are two binding sites on the ribosome, the P (peptidyl or donor site) and the A (aminoacyl) site. At the end of the initiation stage, the fMet-tRNA molecule is empty. In the first step of the elongation cycle, an aminoacyl-tRNA is inserted into the vacant A site on the ribosome. The particular species inserted depends on the mRNA codon that is positioned in the A site. Protein elongation factors and GTP are required for polypeptide chain elongation.
In the next step of the elongation phase, the formylmethionyl residue of the fMet-tRNA located at the peptidyl donor site is released from its linkage to tRNA, and is joined with a peptide bond to the -amino group of the aminoacyl-tRNA in the acceptor site to form a dipeptidyl-tRNA. The enzyme catalyzing this peptide formation is peptidyl transferase, which is part of the 50S ribosomal subunit.
Following the formation of a peptide bond, an uncharged tRNA occupies the P site, whereas a dipeptidyl tRNA occupies the A site. The final phase of the elongation cycle is translocation, catalyzed by elongation factor EF-G and requiring GTP. It consists of three movements:
(2) the movement of fMet-aminoacyl-tRNA from the acceptor site to the peptidyl donor site
(3) the movement or translocation of the ribosome along the mRNA from the 5' toward the 3' terminus by the length of three nucleotides.
The polypeptide chain grows from the amino terminal toward the carboxyl terminal amino acid and remains linked to tRNA and bound to the mRNA-ribosome complex during elongation of the chain. When completed it is released during chain termination. Termination is triggered when a chain termination signal (UAA, UAG, or UGA) is encountered at the A site of the ribosome. Protein release factors bind to the terminator codons triggering hydrolysis by the peptidyl transferase. The polypeptide is released, and the messenger-ribosome-tRNA complex dissociates.
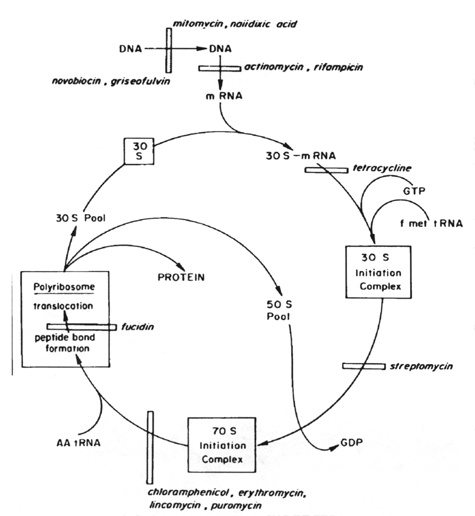
Several medically important antibiotics owe their selective antimicrobial action to a specific attack on the 70S ribosome of bacteria, with mammalian 80S ribosomes left unaffected. Those that act on the 30S ribosome are:
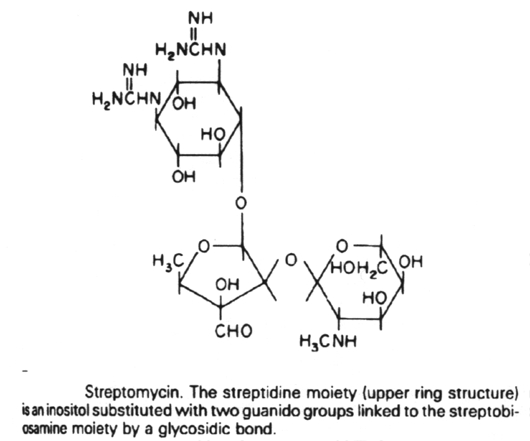
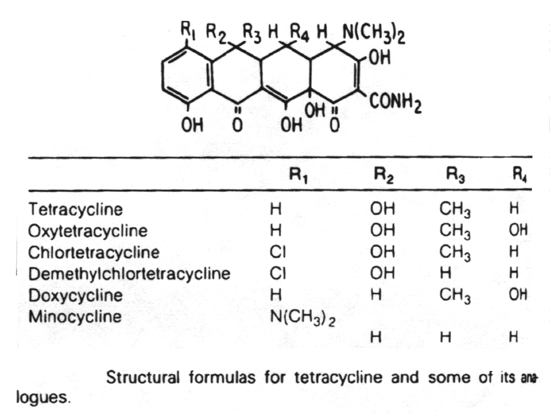
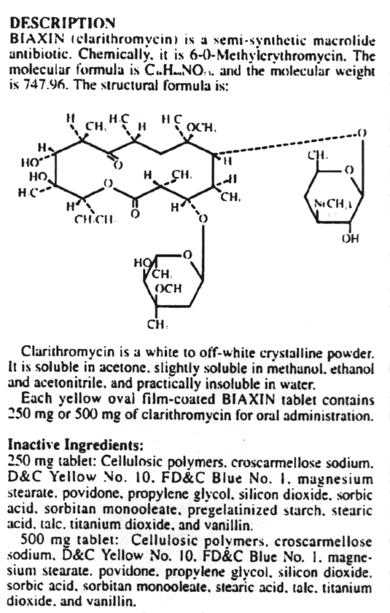
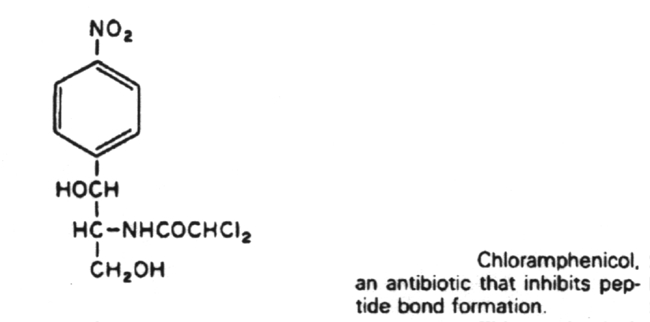
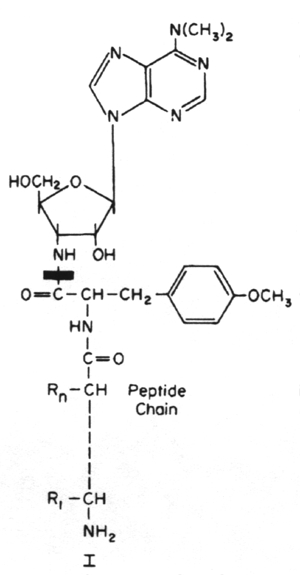
Inhibitors of metabolic pathways via competitive antagonism include:
1. Antibiotics that are active against the cell wall of bacteria include the B-lactams, cycloserine, ethionamide, isoniazid, phosphomycin, vancomycin, bacitracin and ristocetin.
2. Antibiotics that are active against the bacterial cytoplasmic membrane are polymyxin B and E (colistin).
3. Antibiotics that are active against bacterial DNA are the quinolones (nalidixic acid, norfloxacin and ciprofloxacin), which inhibit DNA gyrase, and metronidazole, which fragments DNA.
4. Antibiotics that block transcription in bacteria are actinomycin D and rifampin.
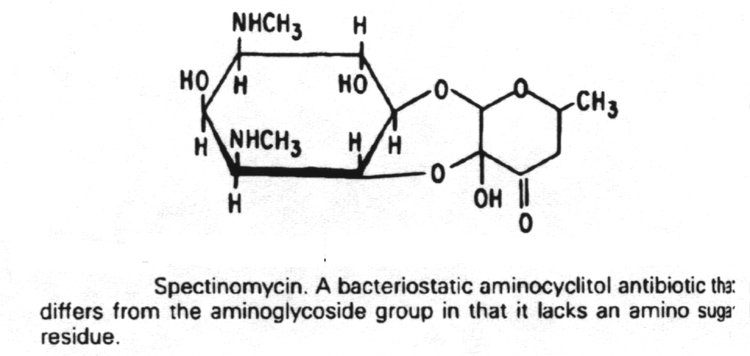
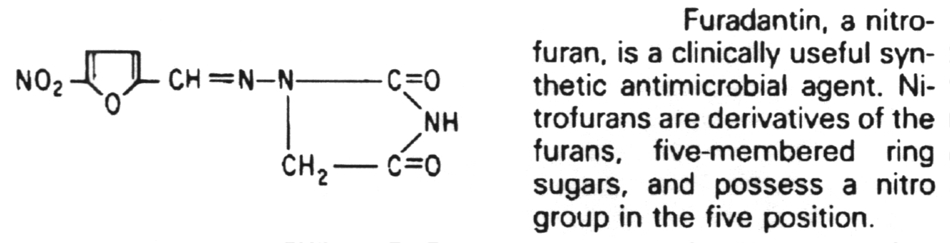
5. Antibiotics that block translation in bacteria by binding to the 30S ribosome are the aminoglycosides, nitrofurans, spectinomycin and the tetracyclines.
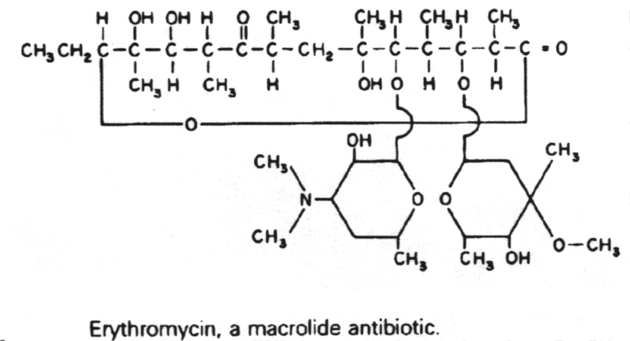
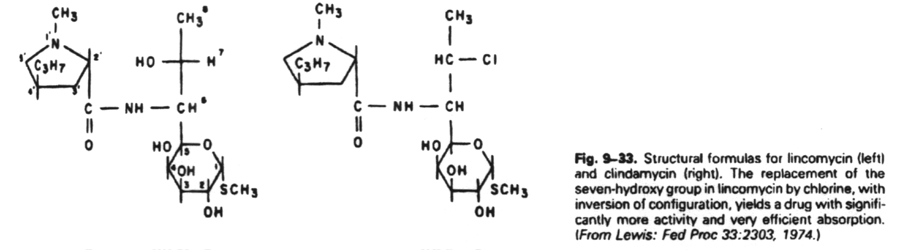
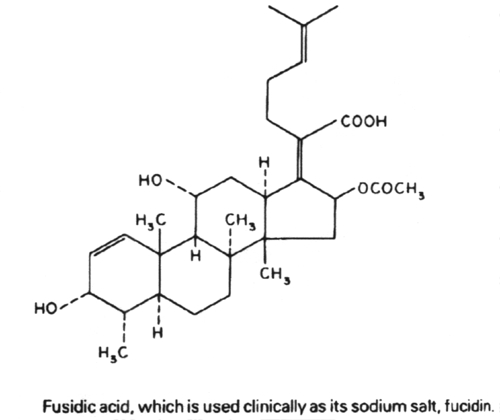
6. Antibiotics that block translation by binding to the 50S ribosome include chloramphenicol, erythromycin, clarithromycin, lincomycin, clindomycin, puromycin and fusidic acid.
7. Antibiotics that act by inhibiting folic acid biosynthesis include the sulfonamides and trimethoprim.
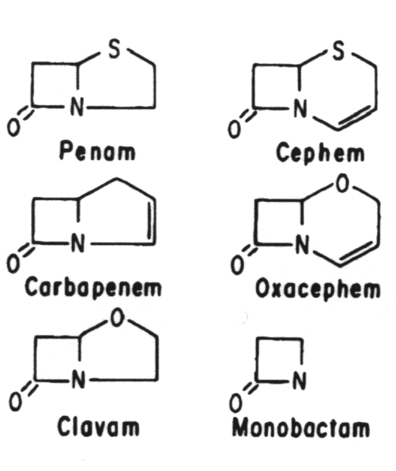
8. The B-lactam antibiotics are related structurally in that they all contain a B-lactam ring. These are the penicillins, cephalosporins, monobactams and thienamycins. They are all analogs of d-alanyl-d-alanine.
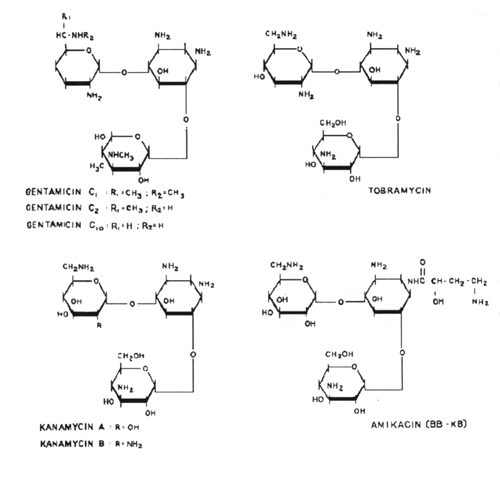
9. The aminoglycoside antibiotics are related structurally in that they all contain a unique aminocyclitol ring structure. These include amikacin, gentamycin, kanamycin, neomycin, streptomycin and tobramycin.
10. The macrolide antibiotics are related structurally in that they all contain a macrocyclic lactone ring of 12-22 carbon atoms, to which one or more sugars are attached. These include erythromycin and clarithromycin
![]()
Previous
Lecture
Top
of lecture
Home
Next
Lecture
![]()